Resources
Insights
Does your High-Throughput Screening workflow demand 2D and 3D Cell Models? Advance your Cardiac Research with the VOLTA Scanner
Case Study
Insights
VOLTA Scanner: A Turnkey, Optical Kinetic Plate Reader for Cardiac Safety Testing
Case Study
Insights
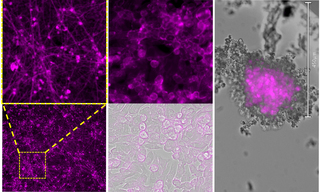
Bursting with Benefits: Optophysiology with BeRST Voltage Sensitive Dye
Case Study
Insights

Lumencor at Photonics West 2025: Advancing Insights with the Power of Light
Video
Insights
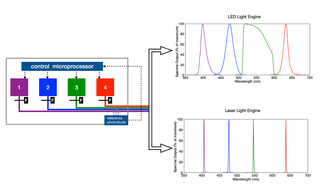
White Paper: Spatial Light Output Characteristics of Solid-State Light Engines
White Paper
Insights
Webinar: Safeguarding the Heart in Drug Discovery
Video
Insights
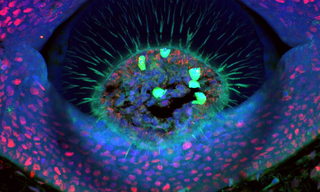
Lumencor’s ZIVA Light Engine for Yokogawa CSU-W1 featured at MBL’s Advanced Research Training Courses
Product News
Insights
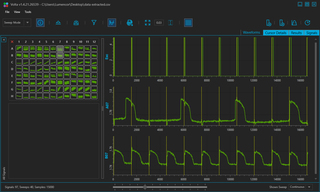
VOLTA Scanner: High-Throughput Cardiomyocyte Electrophysiology with an Optical Pacemaker
Case Study
Insights
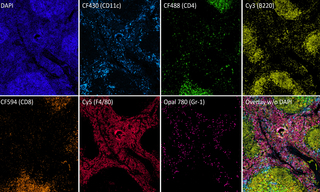
Multiplexed Fluorescence Detection with the new SPECTRA X Light Engine
Case Study
Insights
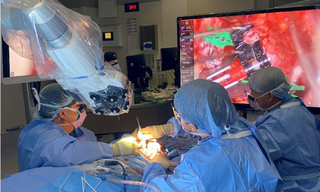
Solid-State Illumination for Intraoperative Imaging with the SPECTRA Light Engine
Case Study
Insights
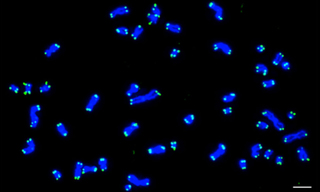
Case Study: Solid-State SOLAs Light Up FISH (Fluorescence In Situ Hybridization)
Case Study
Insights
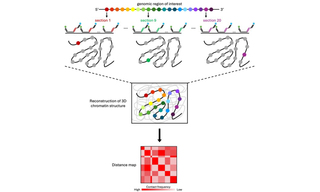
Optical Reconstruction of Chromatin Architecture using the CELESTA Light Engine
Case Study